Introduction
The first version of Cation-Aromatic Database(CAD): Reddy AS, Sastry GM, Sastry GN. Cation-aromatic database. Proteins. 2007 Jun 1;67(4):1179-84.
Cation-Aromatic Database(CAD), is an exhaustive database of cation-aromatic motifs that are present in the experimentally determined 3D structure of proteins, available in the protein data bank (PDB). The database contains statistical data about geometric arrangements, residue preferences, and interaction modes of all the underlying cationic and aromatic moieties, which include cation-σ and cation-π motifs. It aims to advance the knowledge of spatial distributions of cationic and aromatic residues and their interactions in protein structures, as well as how cation-aromatic motifs contribute to protein structure and function. Moreover, CAD has the potential to improve our ability to identify cation-π interactions quickly and facilitate the development of new methods and force fields.
The updated version of CAD (CAD 2.0) includes information of ~27.26 million cation-aromatic motifs identified in the 3D structures of proteins. CAD identifies the position of the cation relative to the arene using three distance parameters r, d1, and d2, and classifies cation-aromatic motifs as either cation-π or cation-σ motifs.
This update of the cation-aromatic database put forth an extensive amount of analysis of the cation-aromatic motifs and their occurrence in different classes of proteins. Among the 27,255,817 cation-aromatic motifs identified in total, 94.09% of the motifs have been observed to be spherical motifs while a mere 5.91% are cylindrical motifs.
The latest version of Cation-Aromatic Database(CAD): Kumar, Y. B., Kumar, N., John, L., Mahanta, H. J., Vaikundamani, S., Nagamani, S., ... & Sastry, G. N. (2023). Analyzing the cation‐aromatic interactions in proteins: Cation‐aromatic database V2. 0. Proteins: Structure, Function, and Bioinformatics.
About Spherical and Cylinderical Motifs
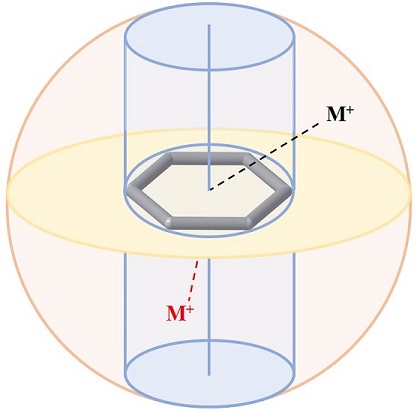
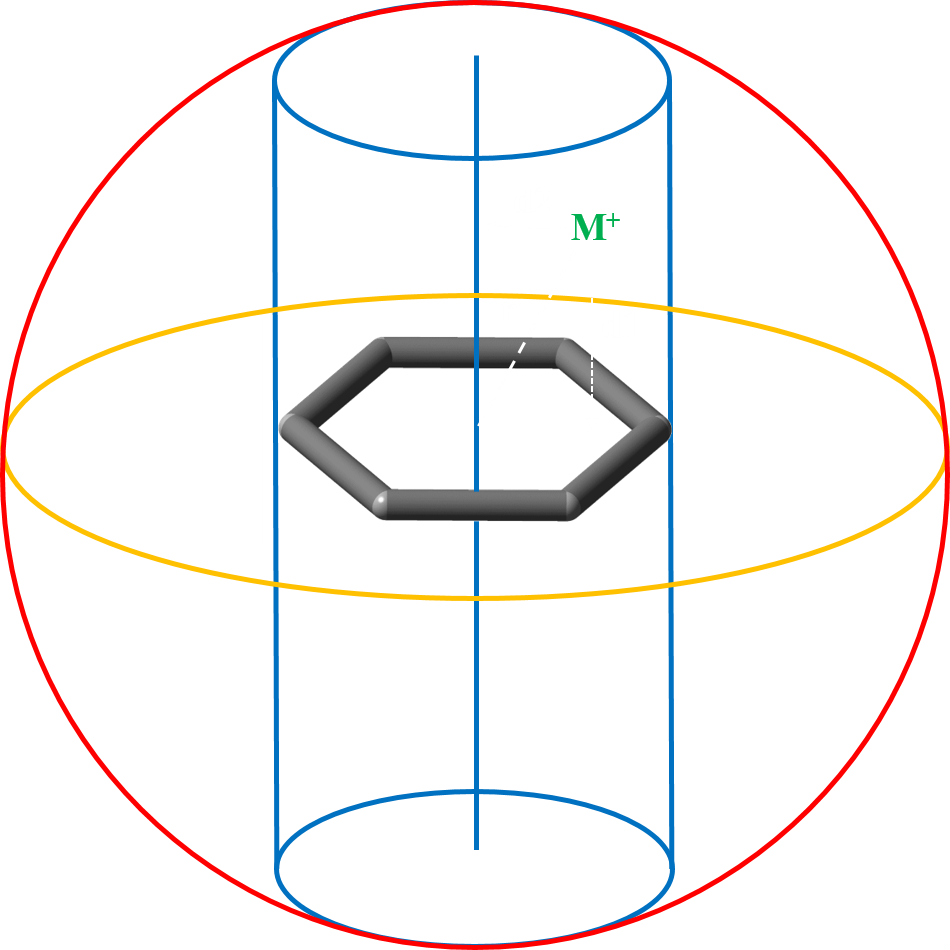
The key feature of CAD is its ability to distinguish between the motifs, where the cation interacts head on with the arene, or interacts along the plane of arene. We have used the distance parameters r, d1, and d2 to identify the relative positions of cationic and aromatic moieties. The distance r is the euclidean distance between the cation and the center of the aromatic residue. The distances d1 and d2 on the other hand are the perpendicular distances from the cation to the plane of the arene and the principal axis of the arene respectively. So, d1 gives information about the strength of interaction while d2 specifies the position of the cation. If the distance d2 is less than the radius of the virtual cylinder, the resultant motif is considered to be a cylindrical motif and undergoes cation-π interaction. On the contrary, if the distance d2 is greater than the radius of the virtual cylinder, it is a spherical motif and takes part in a cation-σ interaction.
All Motifs
ARG-PHE
HIS-PHE
LYS-PHE
ARG-TYR
HIS-TYR
LYS-TYR
ARG-TRP
HIS-TRP
LYS-TRP
ARG-HIS
HIS-HIS
LYS-HIS